What is ZFN-Based Genome Editing?
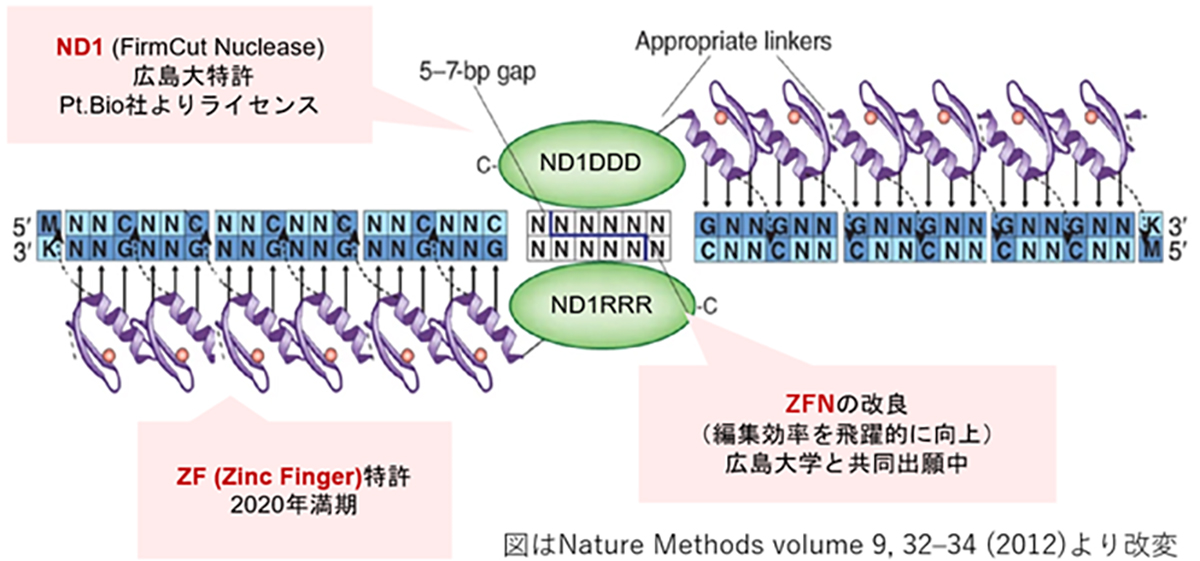
Introduced in 1996, ZFN is the first-generation genome editing enzyme, an artificial protein blending the DNA-binding domain called the Zinc finger motif with the nuclease domain. A single Zinc finger can recognize three base pairs of DNA. By linking multiple Zinc fingers, you can produce a Zinc finger that recognizes extended DNA sequences and binds specifically to target sequences. As endonucleases operate as dimers, when two ZFNs bind to their respective target sequences, a nuclease dimer forms, resulting in DNA cleavage.
Improved ZFN
While ZFNs offer the advantage of a smaller molecular weight, making them easier to load onto vectors like AAV, designing a highly specific ZFN for a target sequence is challenging and time-consuming. However, our joint research with Hiroshima University has achieved high-throughput development of highly specific ZFNs. Leveraging Hiroshima University’s FirmCut nuclease ND1, we established ZFN development technology that achieves a cleavage efficiency comparable to CRISPR-Cas9 in adult retinal cells.
ZFN and Gene Delivery Techniques
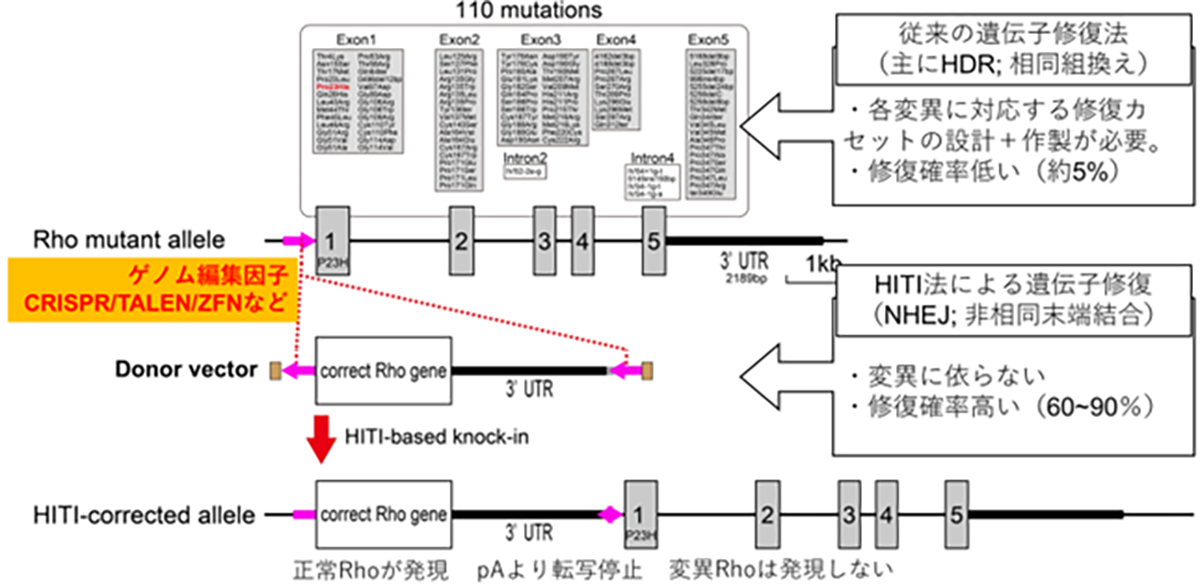
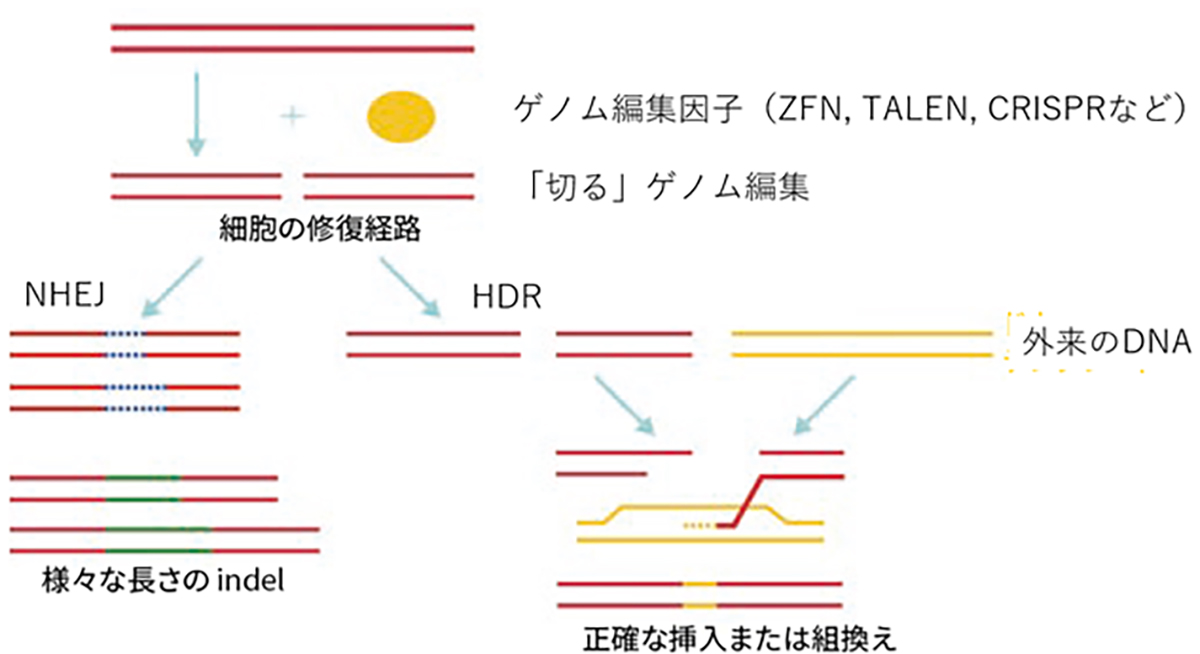
In developing therapeutic agents through HITI gene insertion, it’s vital to craft the right constructs to ensure peak gene functionality, pinpoint the optimal vector for gene delivery, and hone the insertion techniques. Drawing from insights in functional genomics and stem cell development, our research diligently focuses on designing constructs for optimized gene functionality. Collaboratively working with Simprogen Corp. in Kobe, we’re pioneering the development of high-quality adenoviral vectors. Moreover, in partnership with the Kobe Eye Center Hospital, we’re advancing our understanding and methodology of precise gene insertion techniques.